Getting started with Iceberg using Python & DuckDB

Introduction
The purpose of this article is to get you started with Iceberg using Python. We'll start with a quick introduction to Iceberg & table formats in general, then we'll get started with a simple example using Python & DuckDB to query Iceberg tables.

Let's start with the basics
First of all, we need data. One of the most common places to store data is an object storage system like S3 or GCS. If there is some organization to how the files are stored, we can even go as far as calling this a data lake! Having all of our data available in a data lake is a great start, but definitely not enough for modern analytical use cases.
Data Lakes by themselves are great for the storage layer, but they do not provide any consistency guarantees or schema information that we would expect from a more structured data store. To help with this problem, we can use a table format!
What is a table format?
A table format is a specification for how to store data in a collection of files. The files are usually stored in an object store like S3 or GCS. The table format specification defines how to store data in the files, how to store metadata about the files, and how to store metadata about the table itself.
Think of a table format as a protocol for storing data in a collection of files. The files themselves are the same .csv or .parquet files you're used to, but instead of loading them into a data warehouse like Redshift or Snowflake, you define a structure over them using a table format.
Table formats have been around for a long time, but Iceberg and Delta Lake have been gaining popularity lately as two of the most prominent contenders, with Hudi coming in as a close second. The differences between them are very interesting,
but not the focus of this article, maybe a future one!
Why Iceberg?
Iceberg is a table format designed for huge analytic datasets. It offers a clean SQL interface for querying data, full schema evolution, time travel, and rollback, among other killer features.
For an exhaustive comparison of table formats like Iceberg and Delta Lake, check out the benchmark analysis by Brooklyn Data Co.
Now that we have a basic understanding of the tools we'll be using, let's get started!
Getting started with Iceberg using Python
Let's get started! Our goal is to spin up a local Iceberg table using Python and DuckDB. We'll use MinIO as our object storage system, spark to ingest our data, and DuckDB to query the table through the pyiceberg library.
The Data Lake
First of all, we'll need a Data Lake to store our data in. We'll use MinIO, an open-source S3-compatible object storage system. We'll use Docker to run MinIO, but you can also run it locally if you'd like. I've prepared a docker-compose.yml file with all the necessary services for the demo for convenience.
The MinIO section looks like this:
minio:
hostname: "minio"
image: "minio/minio:latest"
container_name: "minio"
ports:
- "9001:9001"
- "9000:9000"
command:
- "server"
- "/data"
- "--console-address"
- ":9001"
volumes:
- "minio:/data"
environment:
MINIO_ROOT_USER: "minio"
MINIO_ROOT_PASSWORD: "minio123"
mc:
depends_on:
- "minio"
image: "minio/mc"
container_name: "mc"
entrypoint: >
/bin/sh -c " until (/usr/bin/mc config host add minio http://minio:9000
minio minio123) do echo "...waiting..." && sleep 1; done; /usr/bin/mc rm
-r --force minio/nyc-taxi-trips; /usr/bin/mc mb minio/nyc-taxi-trips;
/usr/bin/mc policy set public minio/nyc-taxi-trips; exit 0; "
We'll use the `mc` container to create a bucket in MinIO that we can use to store our data. We'll map the `/data/` folder in the MinIO container to a volume so that we can access the data later on. For the demo, I included one `.parquet` file with some data from the NYC Taxi Trips dataset.
The Iceberg table
Now that we have some raw data in our Data Lake, we can start creating our Iceberg table. We'll use the iceberg-spark image to run a small pyspark script that will create the table for us.
The script is basically just two lines of code:
df = spark.read.parquet("/home/iceberg/warehouse/yellow_tripdata_2022-01.parquet")
df.write.saveAsTable("nyctaxi3.trips", format="iceberg")
The first line reads the data from the parquet file and the second line creates the Iceberg table.
To verify that the table was created successfully, we can head over to the MinIO console and see that there is a new folder called `metadata` in the bucket, next to our `data` folder. The `metadata` folder contains all the metadata
about the table, including the schema, the partitioning information, and the location of the data files.
We can even peek into the `<uuid>.metadata.json` file to get an idea of how Iceberg works under the hood.
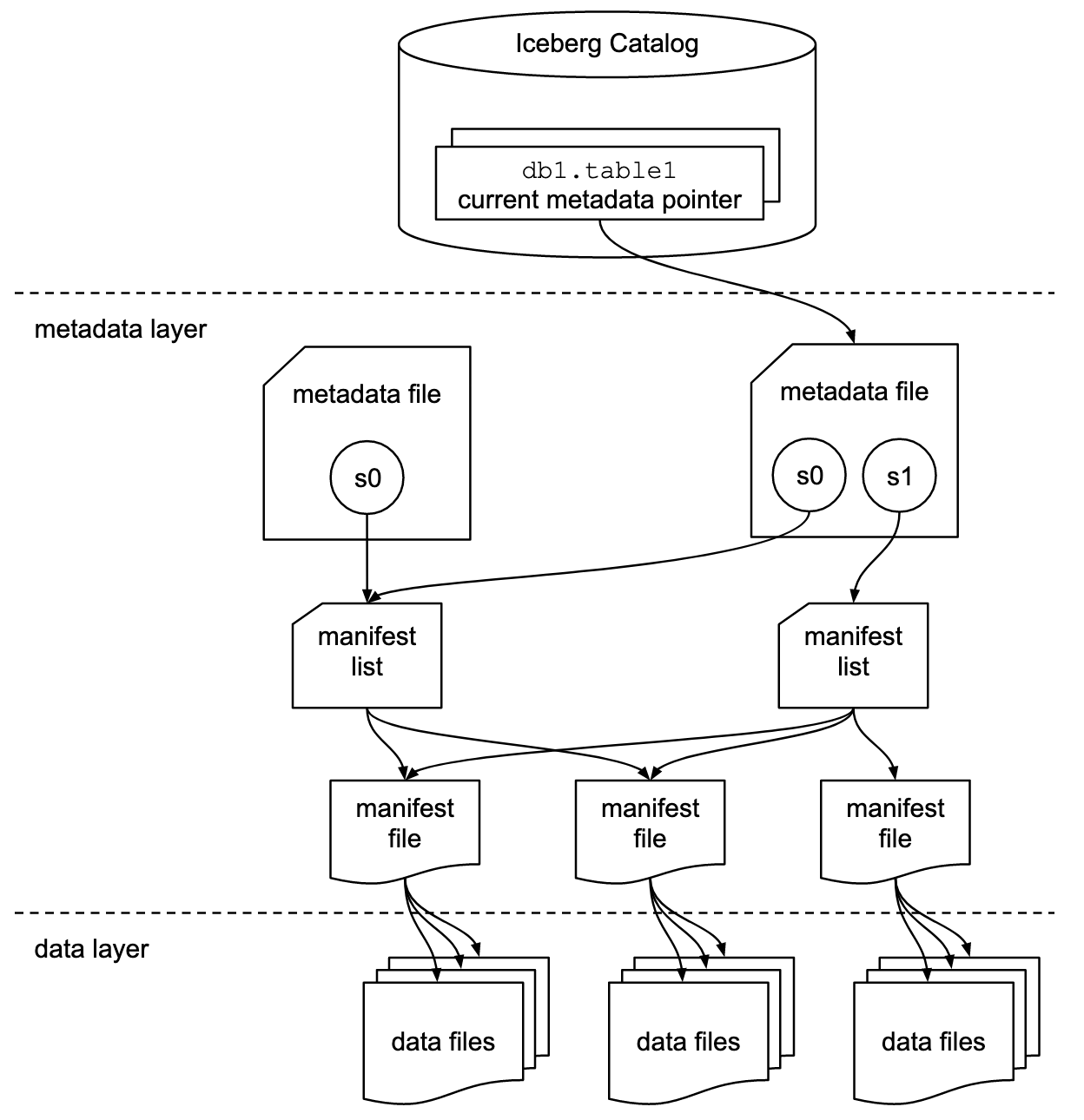
A short explanation of the different parts of an Iceberg table straight from the docs:
Snapshot
A snapshot is the state of a table at some time.
Each snapshot lists all of the data files that make up the table’s contents at the time of the snapshot. Data files are stored across multiple manifest files, and the manifests for a snapshot are listed in a single manifest list file.
Manifest list
A manifest list is a metadata file that lists the manifests that make up a table snapshot. Each manifest file in the manifest list is stored with information about its contents, like partition value ranges, used to speed up metadata operations.
Manifest file
A manifest file is a metadata file that lists a subset of data files that make up a snapshot. Each data file in a manifest is stored with a partition tuple, column-level stats, and summary information used to prune splits during scan planning.
Partition spec
A partition spec is a description of how to partition data in a table. A spec consists of a list of source columns and transforms. A transform produces a partition value from a source value. For example, date(ts) produces the date associated with a timestamp column named ts.
Partition tuple
A partition tuple is a tuple or struct of partition data stored with each data file.
All values in a partition tuple are the same for all rows stored in a data file. Partition tuples are produced by transforming values from row data using a partition spec. Iceberg stores partition values unmodified, unlike Hive tables
that convert values to and from strings in file system paths and keys.
Snapshot log (history table)
The snapshot log is a metadata log of how the table’s current snapshot has changed over time. The log is a list of timestamp and ID pairs: when the current snapshot changed and the snapshot ID the current snapshot was changed to. The snapshot log is stored in table metadata as snapshot-log.
And now, finally,
Time to query!
Alright, now that we got all that out of the way, let’s query the table! Iceberg provides a REST catalog to interact with the tables, which we can use through the pyiceberg library to extract data. The REST catalog is a useful tool that abstracts away the actual catalog implementation.
The relevant `docker-compose.yml` snippet is:
rest:
hostname: "iceberg"
image: "tabulario/iceberg-rest:latest"
container_name: "iceberg"
ports:
- "8181:8181"
environment:
CATALOG_S3_ENDPOINT: "http://minio:9000"
CATALOG_IO__IMPL: "org.apache.iceberg.aws.s3.S3FileIO"
CATALOG_WAREHOUSE: "s3a://nyc-taxi-trips"
AWS_REGION: "us-east-1"
AWS_ACCESS_KEY_ID: "minio"
AWS_SECRET_ACCESS_KEY: "minio123"
The REST catalog is configured to use the S3FileIO implementation, which is the Iceberg implementation for reading and writing data from S3. The S3FileIO implementation requires the AWS credentials to be passed as environment variables, which in our case are the MinIO credentials.
After our REST catalog is up and running, we can use the pyiceberg library to query the table, but first, we need to configure it. This can be easily done by creating a .pyiceberg.yaml file in our home directory with the following
contents:
catalog:
default:
uri: http://0.0.0.0:8181/
s3.endpoint: http://localhost:9000
The uri is the REST catalog endpoint, and the `s3.endpoint` is the S3 endpoint that the REST catalog will use to interact with the S3 bucket.
Now we can finally query the table! We will use the `pyiceberg` library to do so. The most basic example looks like
this:
from pyiceberg.catalog import load_catalog
os.environ["AWS_ACCESS_KEY_ID"] ="minio"
os.environ["AWS_SECRET_ACCESS_KEY"] = "minio123"
CATALOG = load_catalog("default")
table = CATALOG.load_table((namespace, table_name))
print(table.describe())
This will print the table schema so we can verify that all fields are present, with the correct datatypes!
table {
1: VendorID: optional long
2: tpep_pickup_datetime: optional timestamptz
3: tpep_dropoff_datetime: optional timestamptz
4: passenger_count: optional double
5: trip_distance: optional double
6: RatecodeID: optional double
7: store_and_fwd_flag: optional string
8: PULocationID: optional long
9: DOLocationID: optional long
10: payment_type: optional long
11: fare_amount: optional double
12: extra: optional double
13: mta_tax: optional double
14: tip_amount: optional double
15: tolls_amount: optional double
16: improvement_surcharge: optional double
17: total_amount: optional double
18: congestion_surcharge: optional double
19: airport_fee: optional double
}
Looks good, now let's take it a step further by running some analytical queries. By creating a table scan we can easily transform the results into an in-memory DuckDB connection.
con = table.scan().to_duckdb(table_name="trips")
Using this connection we are free to run any SQL query we want! As a bonus, we can convert the results to pandas dataframes with just calling `df()` on the result.
df = con.execute("SELECT * FROM distant_taxi_trips").df()
print(df.head(4))This will print the first 4 rows of the table:
VendorID tpep_pickup_datetime ... congestion_surcharge airport_fee
0 1 2022-01-01 00:35:40+00:00 ... 2.5 0.0
1 1 2022-01-01 00:33:43+00:00 ... 0.0 0.0
2 2 2022-01-01 00:53:21+00:00 ... 0.0 0.0
3 2 2022-01-01 00:25:21+00:00 ... 2.5 0.0
[4 rows x 19 columns]
See how easy that was!? Even with DuckDB, we can easily query the table but converting the results to a pandas dataframe opens up a whole new world of possibilities. We can now use any of the pandas functions to analyze the data or even use it to train a machine learning model!
Conclusion
In this post, we have seen how to use Iceberg to create a table, and how to query it using the REST catalog and the pyiceberg library. We have also seen how to use the pyiceberg library to convert the results of a query into a pandas dataframe.
I hope you can see how easy it is to use Iceberg to create tables and query them. I find that it's not easy to wrap your head around the concepts of Iceberg, especially without playing around with it - but once you do, it's a breeze to use.
Member discussion